Abstract
Introduction: At the time of diagnosis AML the finding of KMT2A gene rearrangement qualifies patient to the group with adverse cytogenetic risk according to ELN 2017. In many hematooncological diseases including AML abnormalities concerning another regions of chromosome 11 are detected. However, their significance for outcome, CR rate, OS is still unknown. The main aim of this study was analysis of chromosome 11 abnormalities in AML patients, focusing on these without KMT2A gene rearrangement and its influence on patients outcome.
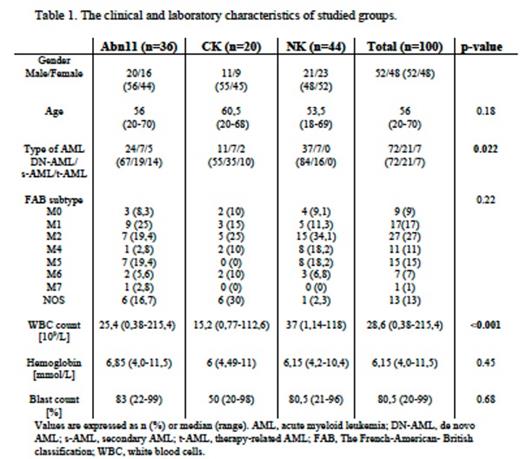
Methods: Karyotype analysis from bone marrow aspirate was performed for patients with chromosome 11 abnormalities (abn11, n=36) and compared to two groups: with complex (CK, n=20) and normal karyotype (NK, n=44). The basic clinical and laboratory data including cytogenetic (karyotype and FISH method) and presence of TP53 deletion were analyzed to characterize in detail the group of patients with chromosome 11 abnormalities.
Patients were treated with intensive chemotherapy, in 33% of them allogeneic cell transplantation (alloHCT) was performed.
Results: In patients with abn11 the most frequent type of aberration was translocation, 11q23(+)/KMT2A(-) and 11q10-25 as a main region involved in aberrations, with minimal narrowing of region to 11q22. In patients with abn11 and translocation (excluding t(9;11)) there was higher frequency occurrence of 1 to 2 additional abnormalities (p=0.033) and lower frequency of complex karyotype existence (p=0.026). The gain of genetic material from chromosome 11 was observed in 33% of patients with abn11 group and the minimal amplified region was 11q22. Furthermore, in patients with amplification of chromosome 11 region, different additional cytogenetic abnormalities (such as monosomy 5 (p=0.045) and 18 (p<0.001)) were found in comparison to patients with other abnormalities of chromosome 11. Additional KMT2Agene copies were often observed in patients with 11q10-25 (p=0.013) and 11pter-qter (p=0.022) region involvement. The lowest frequency of TP53 deletion was in NK group (p=0.002) but comparable in patients with CK and abn11 without impact on OS and CR. The FISH analysis did not revealed KMT2A gene rearrangement in group with normal karyotype. There was particular attention to 11q23 region in patients with AML which enabled abnormality detection in GTG analysis, furthermore KMT2A rearrangement was confirmed or excluded by FISH method. In few cases FISH analysis allowed verification of KMT2A gene status in case of atypical hybrydization pattern occurrence.
Leukocyte counts (p<0.001) and type of AML (de novo, secondary, therapy-related, p=0.022) differs in patients with abn11 in comparison to remained studied subgroups (Table 1.). The lowest blast percentage was observed in patients with 11p rearrangement (p=0.045). There were no significant differences in OS between all 3 groups of patients. CR was achieved in 15% in a CK group and it was significantly lower than in NK and abn11 group (p<0.001). Furthermore alloHCT improve OS in all treated patients (p<0.001). Additional genetic material from chromosome 11 occurrence had negative impact on OS (p<0.001) and CR (p=0.007). Patients with abn11 who achieved CR had lower median age (p=0.01) compared to these without CR. The lowest percentage of CR in abn11 group was observed in patients with additional KMT2A gene copies (p=0.008), 11q10-25 region involvement (p=0.036), HSR (homogeneously staining region) and aneuploidy (p=0.04).
Conclusions: In summary, the most frequent region involved in chromosome 11 aberration was 11q22. Additional material derived from chromosome 11 had negative impact on CR and OS. Cytogenetic and molecular analysis in patients with AML are needed to detect most of abn11. In the era of targeted therapies their identification is very important to qualify the patient for more effective treatment. Both cytogenetic and molecular techniques widespread knowledge about cancer biology, however, classic cytogenetic is still "gold standard" and crucial method in AML diagnosis.
No relevant conflicts of interest to declare.